Blood test detects childhood tumors based on their epigenetic profiles
Share
A new study exploits the characteristic epigenetic signatures of childhood tumors to detect, classify and monitor the disease. The scientists analyzed short fragments of tumor DNA that are circulating in the blood. These "liquid biopsy" analyses exploit the unique epigenetic landscape of bone tumors and do not depend on any genetic alterations, which are rare in childhood cancers. This approach promises to improve personalized diagnostics and, possibly, future therapies of childhood tumors such as Ewing sarcoma. The study has been published in Nature Communications.
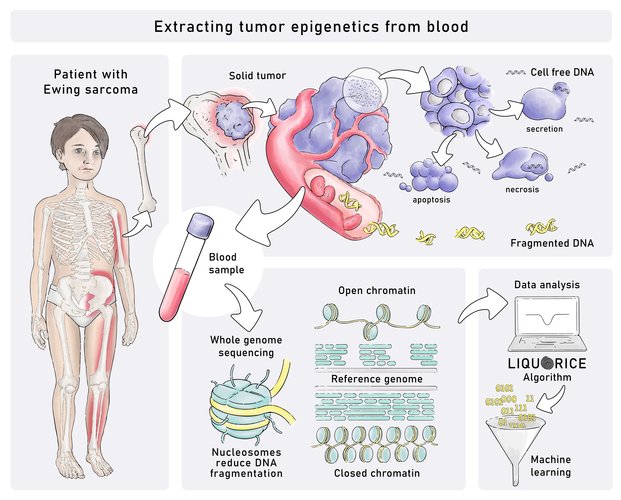
A study led by scientists from St. Anna Children’s Cancer Research Institute (St. Anna CCRI) in collaboration with CeMM Research Center for Molecular Medicine of the Austrian Academy of Sciences provides an innovative method for “liquid biopsy” analysis of childhood tumors. This method exploits the fragmentation patterns of the small DNA fragments that tumors leak into the blood stream, which reflect the unique epigenetic signature of many childhood cancers. Focusing on Ewing sarcoma, a bone tumor of children and young adults with unmet clinical need, the team led by Eleni Tomazou, PhD, St. Anna CCRI, demonstrates the method’s utility for tumor classification and monitoring, which permits close surveillance of cancer therapy without highly invasive tumor biopsies.
In tumors, cancer cells constantly divide, with some of the cancer cells dying in the process. These cells often release their DNA into the blood stream, where it circulates and can be analyzed using genomic methods such as high-throughput DNA sequencing. Such “so-termed liquid biopsy” analyses provide a minimally invasive alternative to conventional tumor biopsies that often require surgery, holding great promise for personalized therapies. For example, it becomes possible to check frequently for molecular changes in the tumor. However, the use of liquid biopsy for childhood cancers has so far been hampered by the fact that many childhood tumors have few genetic alterations that are detectable in DNA isolated from blood plasma.
Exploiting tumor-specific epigenetic patterns
Cell-free DNA from dying tumor cells circulates in the blood in the form of small fragments. Their size is neither random nor determined solely by the DNA sequence. Rather, it reflects how the DNA is packaged inside the cancer cells, and it is influenced by the chromatin (i.e., complex of DNA, protein and RNA) structure and epigenetic profiles of these cells. This is very relevant because epigenetic patterns – sometimes referred to as the “second code” of the genome – are characteristically different for different cell types in the human body. Epigenetic mechanisms lead to changes in gene function that are not based on changes in the DNA sequence but are passed on to daughter cells. The analysis of cell-free DNA fragmentation patterns provides a unique opportunity to learn about the epigenetic regulation inside the tumor without having to surgically remove tumor cells or even know whether and where in the body a tumor exists.
“We previously identified unique epigenetic signatures of Ewing sarcoma. We reasoned that these characteristic epigenetic signatures should be preserved in the fragmentation patterns of tumor-derived DNA circulating in the blood. This would provide us with a much-needed marker for early diagnosis and tumor classification using the liquid biopsy concept”, explains Dr. Tomazou, Principal Investigator of the Epigenome-based precision medicine group at St. Anna CCRI.
Machine learning increases sensitivity
The new study benchmarks various metrics for analyzing cell-free DNA fragmentation, and it introduces the LIQUORICE algorithm for detecting circulating tumor DNA based on cancer-specific chromatin signatures. The scientists used machine-learning classifiers to distinguish between patients with cancer and healthy individuals, and between different types of pediatric sarcomas. “By feeding these machine learning algorithms with our extensive whole genome sequencing data of tumor-derived DNA in the blood stream, the analysis becomes highly sensitive and in many instances outperforms conventional genetic analyses”, says Dr. Tomazou.
When asked about potential applications, she explains: “Our assay works well, we are very excited. However, further validation will be needed before it can become part of routine clinical diagnostics.” According to the scientists, their approach could be used for minimally invasive diagnosis and, but also as a prognostic marker, monitoring which patient responds to therapy. Additionally, it might serve as a predictive marker during neoadjuvant therapy (i.e., chemotherapy before surgery), potentially enabling dose adjustments according to treatment response. “Right now, most patients receive very high doses of chemotherapy, while some patients may be cured already with a less severe therapy, which would reduce their risk of getting other cancers later in life. There is a real medical need for adaptive clinical trials and personalized treatment of bone tumors in children.”
International scientific collaboration
This study was led by St. Anna CCRI in collaboration with CeMM Research Center for Molecular Medicine of the Austrian Academy of Sciences, Ludwig Boltzmann Institute for Rare and Undiagnosed Diseases, Medical University of Vienna, collaborating with multiple institutions in Austria, Germany, Norway, and France.
About epigenetics
Epigenetics is the link between genes and their environment. It contributes to gene regulation and controls which genes are active or inactive at specific time points. Epigenetic mechanisms lead to changes in gene function that are not based on changes in the DNA sequence – for example through mutation – but are passed on to daughter cells. Since childhood cancers often harbor few genetic alterations, their epigenetic patterns are promising markers for non-invasive diagnostics using liquid biopsy.
Publication:
"Multimodal analysis of cell-free DNA whole genome sequencing for pediatric cancers with low mutational burden" was published on Nature Communications on May 28, 2021. Doi: https://doi.org/10.1038/s41467-021-23445-w
Authors:
Peter Peneder*, Adrian M Stütz*, Didier Surdez, Manuela Krumbholz, Sabine Semper, Mathieu Chicard, Nathan C Sheffield, Gaelle Pierron, Eve Lapouble, Marcus Tötzl, Bekir Erguner, Daniele Barreca, Andre F Rendeiro, Abbas Agaimy, Heidrun Boztug, Gernot Engstler, Michael Dworzak, Marie Bernkopf, Sabine Taschner-Mandl, Inge M Ambros, Ola Myklebost, Perrine Marec-Bérard, Susan Ann Burchill, Bernadette Brennan, Sandra J Strauss, Jeremy Whelan, Gudrun Schleiermacher, Christiane Schaefer, Uta Dirksen, Caroline Hutter, Kjetil Boye, Peter F Ambros, Olivier Delattre, Markus Metzler, Christoph Bock#, Eleni M Tomazou#
*Shared first authors
#Corresponding authors
Funding:
This study was funded by the Austrian National Bank’s Jubilaumsfonds (OeNB), a charitable donation from Kapsch Group and the Austrian Science Fund (FWF).